Blog Post
LLMs in healthcare: tackling implementation challenges methodically
Color

In our previous post, we delved into the multifaceted challenges faced by healthcare providers in the realm of cancer care: overburdened clinicians, patients navigating a fragmented system, and the constant struggle to stay updated with ever-evolving medical guidelines. We discussed how large language models (LLMs) can assist by quickly extracting meaningful data from medical records, interpreting complex guidelines, and generating patient-specific recommendations.
This is a challenging set of capabilities to build in traditional software applications. Patient charts can come in a variety of formats based on the EHR used and often critical information can be found in unstructured fields like the clinician’s notes from a visit. Similarly, clinical guidelines are written for clinicians, and translating them into executable code involves developers interpreting clinical language, which is error-prone, and a process that needs to be repeated whenever new guidelines are published. While a traditional software approach could address these issues, it’s too tedious and costly to be tackled.
Enter LLMs. LLMs are advanced computational tools that excel at extracting and reasoning about information embedded in text, which makes them particularly suited to addressing this problem. By harnessing the power of LLMs, we unlock our ability to solve complex tasks like the ones described above. This not only streamlines complex tasks but also opens new pathways for innovation and improvement in cancer care.
The Challenge with LLMs
Despite the groundbreaking capabilities that LLMs possess, they still face certain challenges. As we explored ways to tackle the problem space we were delving into, we encountered a few hurdles:
- Extracting meaning from data formats that encode information in non-standard or uncommon structures: In clinical guidelines, it’s common to find clinician workflows and decision criteria presented as flow charts. Moreover, these charts are typically included as images in the documents, which adds another layer of complexity to extracting the flow of logic they contain. LLMs are not natively adept at understanding information presented in unusual formats. This necessitates advanced techniques for image recognition and logic interpretation to accurately parse and utilize the embedded information.
- Evaluating the accuracy of the model’s output: Ensuring that the results produced by the LLM are accurate and reliable is a significant challenge. We need to define validation techniques and cross-reference with an established and reliable data source to verify the correctness of the information generated by the model.
- Aligning the output that LLMs give us to match what we need: Often, the responses generated by LLMs need to be fine-tuned to fit specific requirements or contexts. This involves adjusting prompts, employing post-processing techniques, and sometimes integrating additional layers of logic to ensure the output meets our exact needs.
- Interpretability of the output: Understanding how and why an LLM arrived at a particular output is crucial. If the LLMs predictions are off, or if they are not rooted in the patient record or clinical guidelines, we risk losing the trust of those using it. Developing methods to interpret and explain the model’s reasoning is essential for building and maintaining trust in its use.
As we worked to address these hurdles, we found ourselves navigating a noisy landscape full of hammers desperately seeking nails. To RAG or not to RAG? Should we throw RLHF at it? Maybe fine-tune the model? Try few-shot learning? Or perhaps some careful prompt engineering? Each approach had its own merits, and we had to choose the best one/combination for the task at hand.
Rising to the Challenge: A Systematic Approach
Narrow scope to gain focus
To get the clarity we needed, we anchored around our goal of developing a tool that significantly enhances the quality of care for cancer patients while easing the burden on healthcare providers. We decided to start by focusing on a single task for a specific type of cancer and executing it exceptionally well. Ultimately, we chose to concentrate on generating workup plans for breast cancer patients.
In the context of a primary care clinic, an emergency room, or pre-scheduling of an oncology appointment, a tool that can assist a non-specialist in ordering the right workup for a patient needs to:
- Understand a patient’s record holistically,
- Search through trusted guidelines and identify the recommended workup for the patient,
- Identify which labs, imaging, biopsies, etc. are recommended but not yet completed and surface them,
- Provide the clinical user with a clear explanation of its recommendations, their relevance to the patient, and link back to the guidelines for each one.
Focusing on this specific problem allowed us to refine our interaction with clinical guidelines, enabling faster development and validation of our value hypothesis. By narrowing our focus to generating workup plans for breast cancer patients, we built a knowledge base grounded in widely trusted guidelines like NCCN, USPSTF, and UpToDate. This knowledge base details every possible workup and its relevance, written in the language of clinicians. This preprocessing step helps bypass the challenge of working with data formats that LLMs struggle to process natively.
Defining Success
With this narrowed scope, we needed to decide how to start utilizing AI and evaluate the system’s quality. Our primary concern was the outcome rather than the specific techniques employed. Extensive research demonstrated the efficacy of using GPT-4 in medical settings with in-context tuning techniques, indicating that a system could be built using these methods. Therefore, we focused on determining the best architectural decisions and the optimal combination of in-context tuning techniques to ensure the highest performance. To guide these decisions, we defined clear success criteria for the system:
- Accuracy: Ensuring the generated workup plans are precise and reliable.
- Interpretability: Providing clear explanations and citations for recommended workups, grounded in information from the patient record and our knowledge base.
- Reproducibility: Ensuring that the system consistently produces the same output when given the same input, thereby validating the reliability of its performance.
- Generalizability: Ensuring that the methods and data used during the development of the system are representative of its performance on new, unseen data, thereby confirming its effectiveness in diverse and real-world scenarios.
These criteria defined two key aspects of our evaluation:
- The dimensions along which we measured performance.
- A performance requirement for each dimension was established to assess the system’s production readiness, ensuring it could be launched within our product.
Measuring Success
To measure and evaluate our system’s performance along the defined dimensions, we needed a representative set of de-identified patient cases that we could evaluate our performance against. This allows us to directly compare the system’s recommendations with those of the experts.
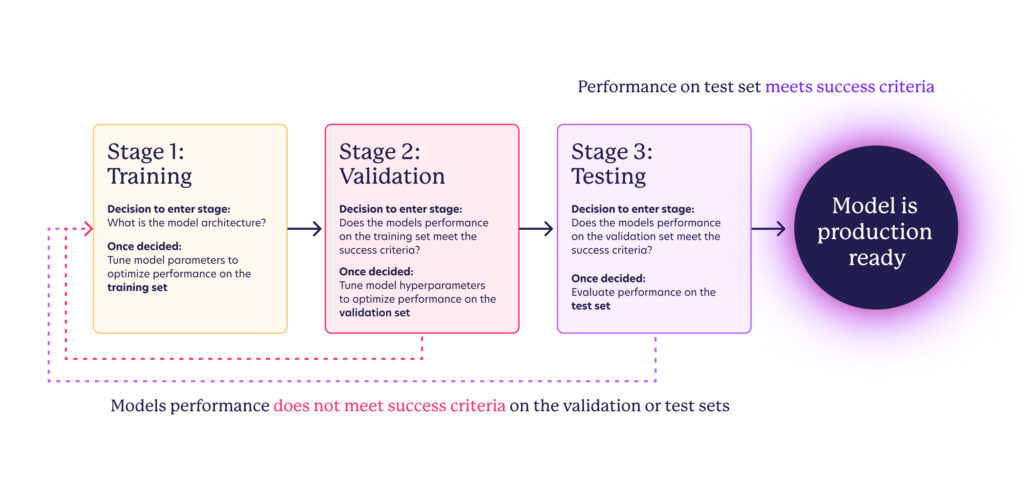
In traditional machine learning model development, three datasets of distinct examples are used:
- Training Dataset: Used to train the model by tuning its parameters.
- Validation Dataset: Used to tune the hyperparameters and to select the best model. This dataset helps to fine-tune the model to improve its performance.
- Test Dataset: Used to evaluate the final model’s performance. This dataset is only used once the model has been trained and validated, and it is meant to provide an unbiased assessment of the model’s performance.
With those datasets, the stages of model development follow:
Since we are using GPT-4o, a pre-trained state-of-the-art generalist LLM, we could skip the initial training stage and the need for a training set, focusing instead on curating validation and test sets. The model development flow is adapted to:
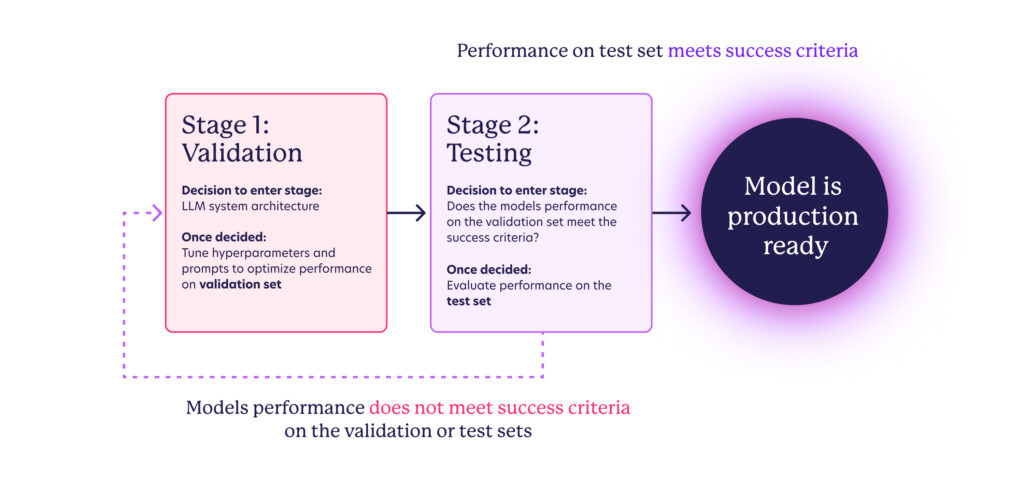
The intended outcome of this development loop is to create a system whose behavior is well understood and aligned with our product needs. Capturing the degree of alignment numerically involves defining and selecting metrics for the success criteria outlined in the previous section. However, the true assessment of this alignment ultimately depends on the volume and quality of data in the validation and test sets. This is where cross-functional collaboration plays a key role – in order to trust that the metrics observed during testing accurately predict real-world performance, we must use scenarios that reflect actual patient data. Color’s clinical experts, along with external advisors, have invested considerable effort in synthesizing test patients that cover various scenarios we might encounter, including the expected recommendations for each one.
Tip: When building an AI assistant for a specialized task, work very closely with your experts on curating the data for tuning and evaluating the system.
Putting it all together
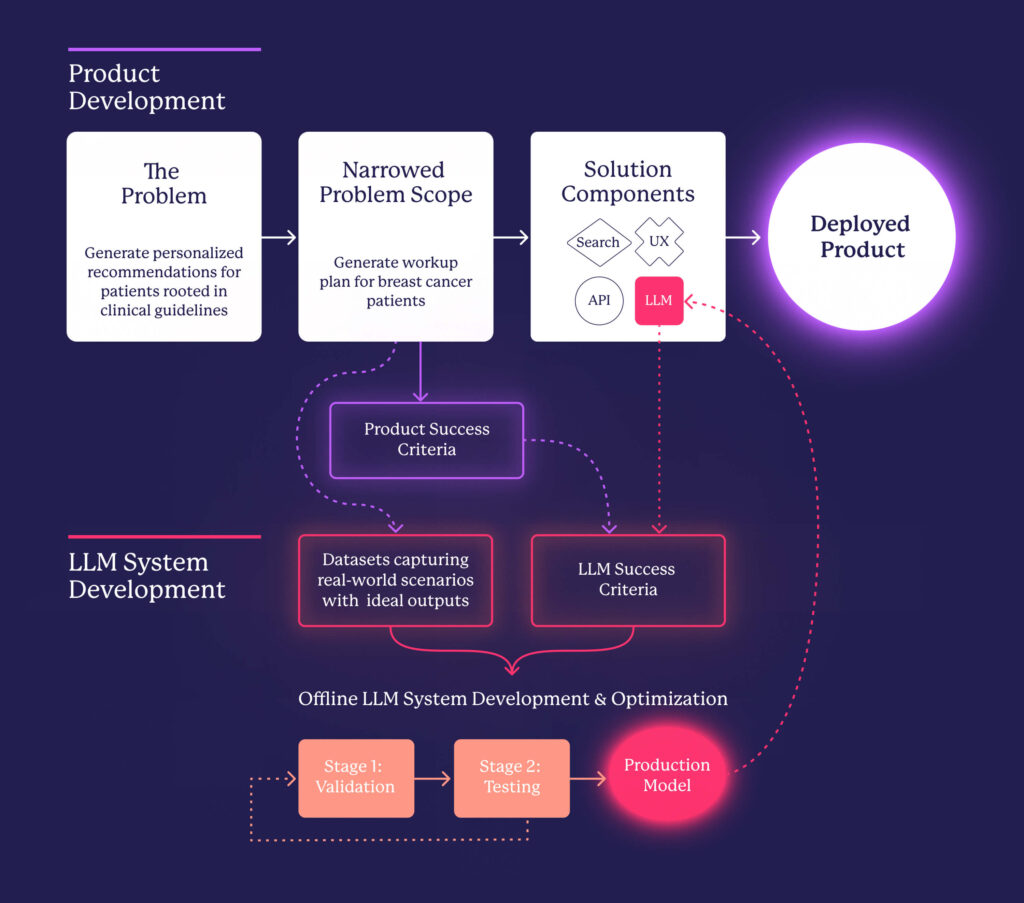
We have demonstrated how leveraging LLMs can significantly enhance the identification and addressing of challenges faced by healthcare providers. While these models offer remarkable capabilities, they also introduce a distinct set of complexities. By methodically navigating these challenges—ranging from extracting meaningful data to ensuring accuracy, interpretability, and alignment of outputs—we have developed a systematic approach that enables the creation of reliable, AI-driven solutions. The following diagram illustrates our process, from defining the problem to narrowing the scope and ultimately integrating LLMs to create a product that effectively addresses a real-world need.
Next Up
In our next post, we’ll go into detail about how we defined metrics to capture the dimensions along which we evaluated our system and the experimentation framework and tooling that we put in place to be able to iterate quickly.